Featured research articles (work led or co-led by our lab)
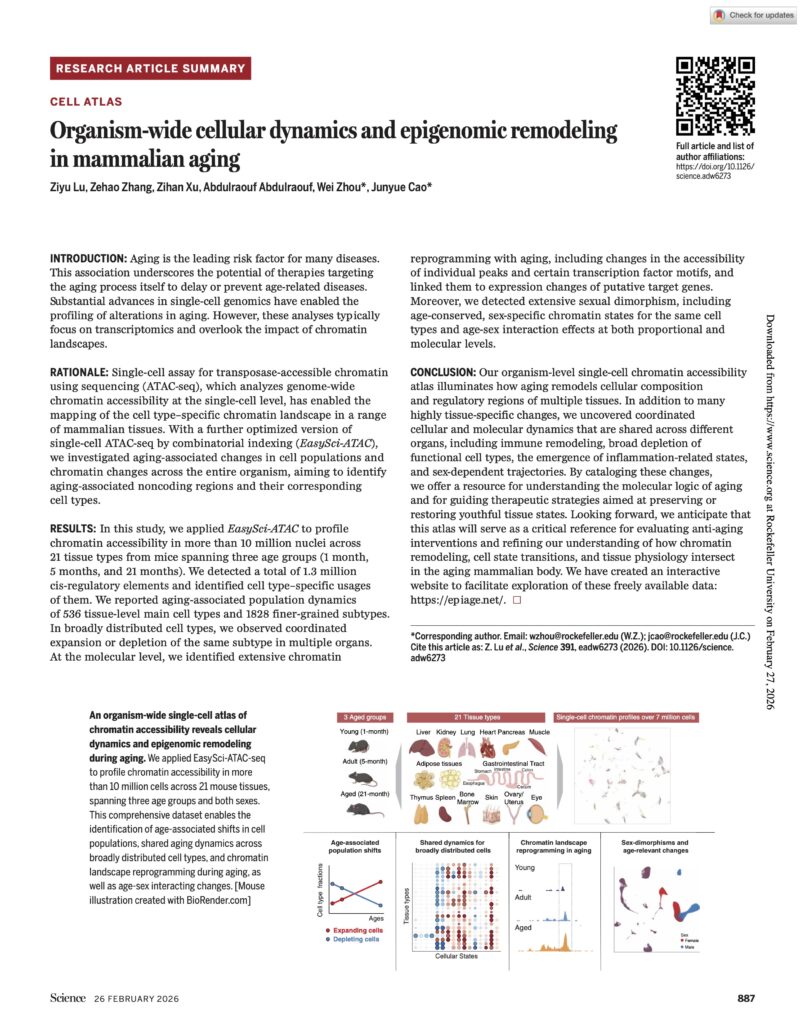
Lu Z, et al. Organism-wide cellular dynamics and epigenomic remodeling in mammalian aging .
Science (2026)
Liao A., et al. Transcript-guided targeted cell enrichment for scalable single-nucleus RNA sequencing
Cell Genomics (2025)
Xu, Z. et al. Uncovering Convergent Cell State Dynamics Across Divergent Genetic Perturbations Through Single-Cell High-Content CRISPR Screening.
BioRxiv preprint (2025)
Abdulraouf, Jiang, et al. Optics-free Spatial Genomics for Mapping Mouse Brain Aging.
BioRxiv preprint (2024)
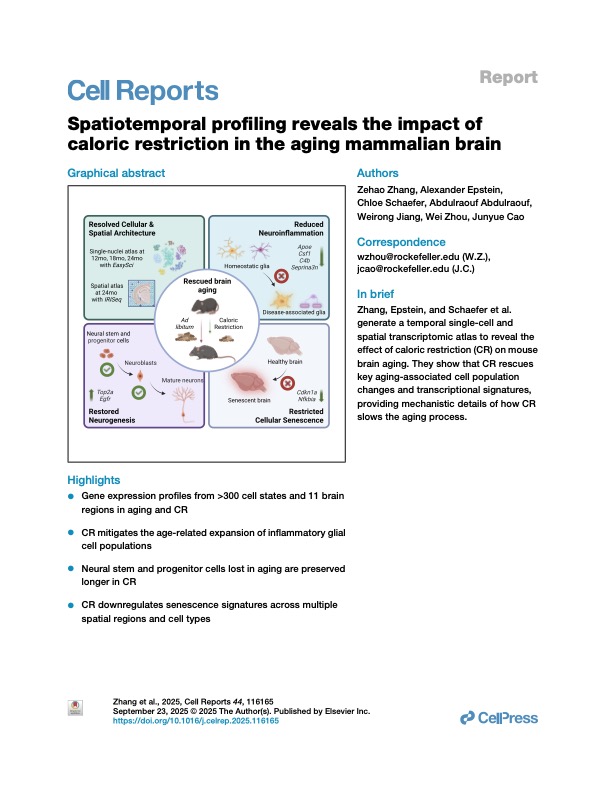
Zhang, Epstein, Schaefer et al. Spatiotemporal profiling reveals the impact of caloric restriction in the aging mammalian brain
Cell Reports (2025)
Zhou W., Cao J. The Genomics of Aging at the Single-Cell Scale. (Review Article).
Annual Review of Genomics and Human Genetics (2025)
Zhang, Z. et al. A Panoramic View of Cell Population Dynamics in Mammalian Aging.
Science (2024)
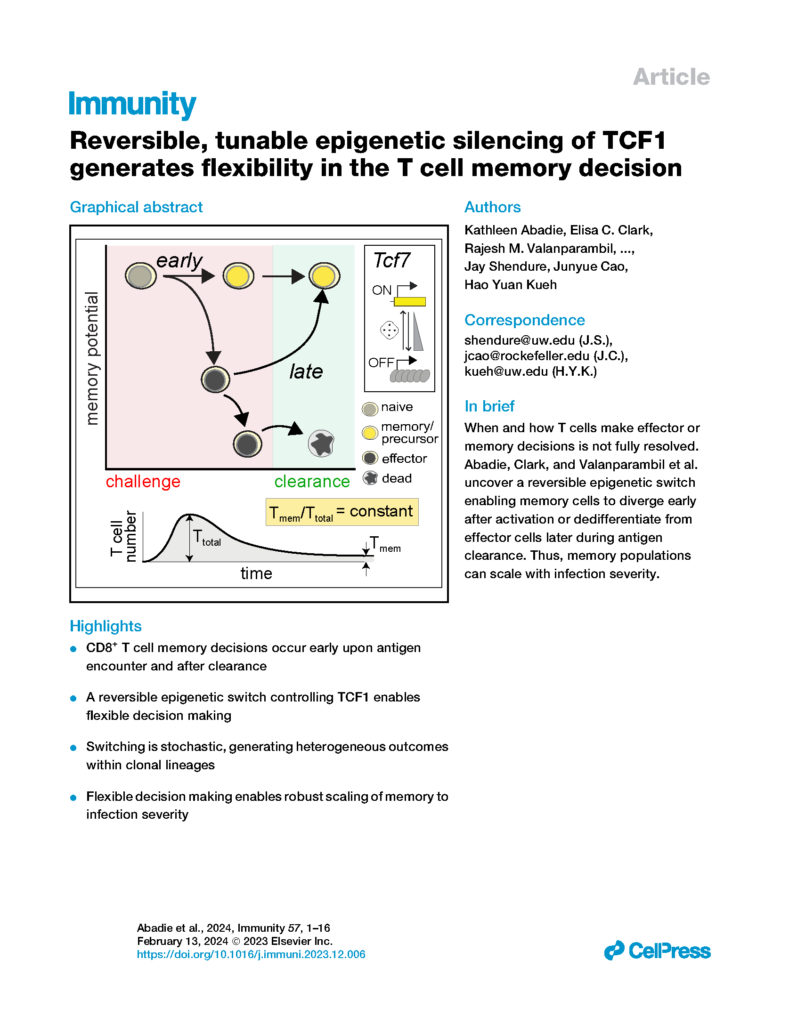
Abadie, Clark, Valanparambil, et al. Reversible, tunable epigenetic silencing of TCF1 generates flexibility in the T cell memory decision. (with Kueh and Shendure Labs)
Immunity (2024)
Sziraki, Lu, Lee, et al. A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in human and mouse brains.
Nature Genetics (2023)
Huang, Henck, Qiu et al. Single-cell, whole embryo phenotyping of mammalian developmental disorders. (with Spielmann and Shendure Labs)
Nature (2023)
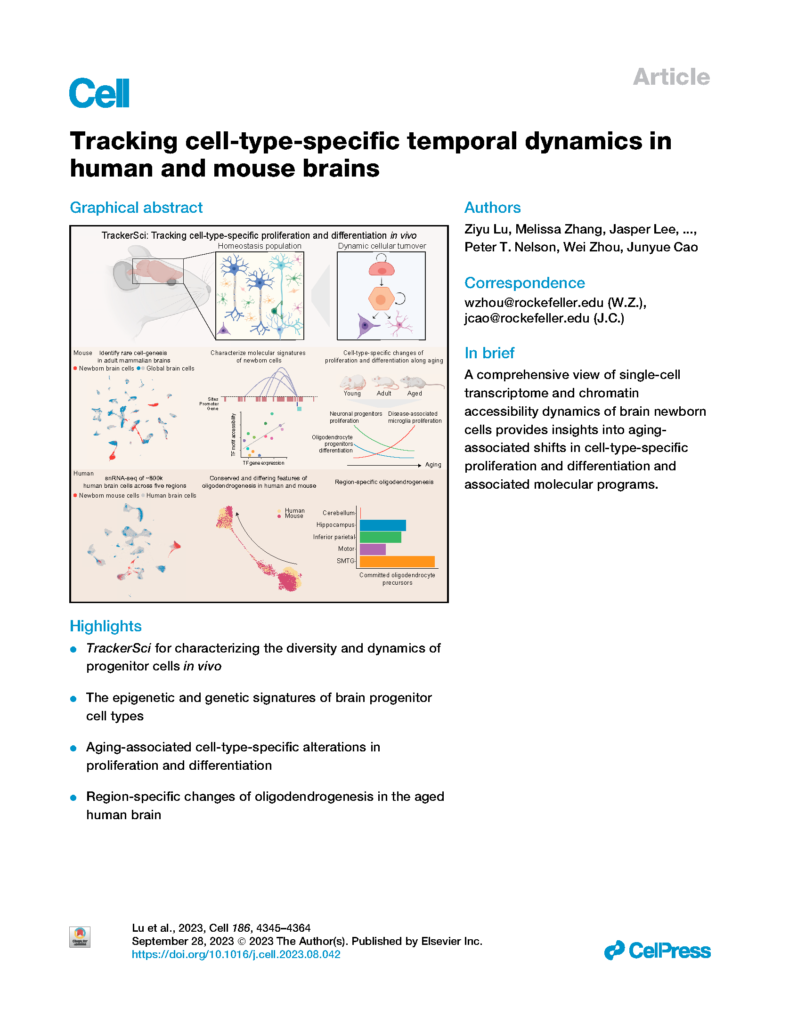
Lu, Zhang, et al. Tracking cell-type-specific temporal dynamics in human and mouse brains.
Cell (2023)
Xu, et al. Dissecting key regulators of transcriptome kinetics through scalable single-cell RNA profiling of pooled CRISPR screens.
Nature Biotechnology (2023)
Zhou, Melamed, Banyai, et al. Expanding the binding specificity for RNA recognition by a PUF domain. (With Fields lab)
Nature Communications (2021)
All Publications
* denotes co-first authorship; # denotes co-corresponding authorship
2026
- Lu Z., Zhang Z., Xu Z., Abdulraouf A., Zhou W.#, & Cao J#. Organism-wide cellular dynamics and epigenomic remodeling in mammalian aging. Science (2026). DOI: 10.1126/science.adw6273
2025
- Liao A., Zhang Z., Sziraki A., Abdulraouf A., Xu Z., Lu Z., Zhou W.#, & Cao J#. Transcript-guided targeted cell enrichment for scalable single-nucleus RNA sequencing. Cell Genomics (2025) DOI: 10.1016/j.xgen.2025.101101
- Xu Z#., Lu Z., Ugurbil A., Abdulraouf A., Liao A., Zhang J, Zhou W.#, & Cao J#. Uncovering Convergent Cell State Dynamics Across Divergent Genetic Perturbations Through Single-Cell High-Content CRISPR Screening. BioRxiv (2025) 10.1101/2025.05.02.651939
- Zhang Z*., Epstein A*., Schaefer A*., Abdulraouf A., Jiang W., Zhou W#., Cao J.#. Spatiotemporal profiling reveals the impact of caloric restriction on mammalian brain aging. Cell Reports (2025). DOI: 10.1016/j.celrep.2025.116165
- Zhou, W., & Cao, J#. The Genomics of Aging at the Single-Cell. Annual Review of Genomics and Human Genetics (2025) DOI: 10.1146/annurev-genom-120523-024422
2024
- Abdulraouf, A*., Jiang, W*., Xu, Z., Zhang, Z., Isakov, S., Raihan, T., Zhou, W.#, & Cao, J#. (2024). Optics-free Spatial Genomics for Mapping Mouse Brain Aging. BioRxiv (2024) https://doi.org/10.1101/2024.08.06.606712
- Zhang, Z., Schaefer, C., Jiang, W., Lu, Z., Lee, J., Sziraki, A., Abdulraouf, A., Wick, B., Haeussler, M., Li, Z., Molla, G., Satija, R., Zhou, W.#, & Cao, J#. ” A Panoramic View of Cell Population Dynamics in Mammalian Aging” Science (2024) DOI: 10.1126/science.adn39
- Qiu, C*., Martin, B. K.*, Welsh, I. C*., Daza, R. M., Le, T.-M., Huang, X., Nichols, E. K., Taylor, M. L., Fulton, O., O’Day, D. R., Gomes, A. R., Ilcisin, S., Srivatsan, S., Deng, X., Disteche, C. M., Noble, W. S., Hamazaki, N., Moens, C. B., Kimelman, D., … Shendure, J. “A single-cell time-lapse of mouse prenatal development from gastrula to birth”. Nature (2024) 626 (8001): 1084–93.
- Abadie, K., * Clark, E. C., * Valanparambil, R. M., * Ukogu, O., Yang, W., Daza, R. M., Ng, K. K. H., Fathima, J., Wang, A. L., Lee, J., Nasti, T. H., Bhandoola, A., Nourmohammad, A., Ahmed, R., Shendure, J., # Cao, J., # & Kueh, H. Y#. “Reversible, tunable epigenetic silencing of TCF1 generates flexibility in the T cell memory decision” Immunity (2024) doi: 10.1016/j.immuni.2023.12.006
2023
- Sziraki, A.,* Lu, Z.,* Lee, J.,* Banyai, G., Anderson, S., Abdulraouf, A., Metzner, E., Liao, A., Epstein, A., Schaefer C., Xu, Z., Zhang, Z., Gan, L., Nelson, P. T., Zhou, W.,# Cao, J.# A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in human and mouse brains. Nature Genetics (2023) https://doi.org/10.1038/s41588-023-01572-y
- Huang, X.,* Henck, J.,* Qiu, C.,* Sreenivasan, V. K. A., Balachandran, S., Behncke, R., Chan, W.-L., Despang, A., Dickel, D. E., Haag, N., Hägerling, R., Hansmeier, N., Hennig, F., Marshall, C., Rajderkar, S., Ringel, A., Robson, M., Saunders, L., Srivatsan, S. R., Ulferts,S., Wittler, L., Zhu, Y., Kalscheuer, V., Ibrahim, D., Kurth, I., Kornak, U., Beier, D., Visel, A., Pennacchio, L., Trapnell, C., Cao, J.,# Shendure, J.,# Spielmann, M#. Single-cell, whole-embryo phenotyping of mammalian developmental disorders. Nature (2023). https://doi.org/10.1038/s41586-023-06548-w
- Sziraki, A., Zhong, Y., Neltner, A. M., Niedowicz, D., Rogers, C. B., Wilcock, D. M., Nehra, G., Neltner, J. H., Smith, R. R., Hartz, A. M., Cao, J., & Nelson, P. T. A high-throughput single-cell RNA expression profiling method identifies human pericyte markers. Neuropathology and Applied Neurobiology (2023), e12942.
- Lu, Z.,* Zhang, M.,* Lee, J., Sziraki, A., Anderson, S., Zhang, Z., Xu, Z., Jiang, W., Ge, S., Nelson, P., Zhou, W.#, Cao, J#. Tracking cell-type-specific temporal dynamics in human and mouse brains. Cell 186, 4345–4364.e24 (2023). doi.org/10.1016/j.cell.2023.08.042
- Xu, Z., Sziraki, A., Lee, J., Zhou, W#. & Cao, J#. Dissecting key regulators of transcriptome kinetics through scalable single-cell RNA profiling of pooled CRISPR screens. Nature Biotechnology (2023) doi:10.1038/s41587-023-01948-9.
- Kim, J., Lee, J., Li, X., Lee, H. S., Kim, K., Chaparala, V., Murphy, W., Zhou, W., Cao, J., Lowes, M. A., & Krueger, J. G. Single-cell transcriptomics suggest distinct upstream drivers of IL-17A/F in hidradenitis versus psoriasis. The Journal of Allergy and Clinical Immunology (2023), 152(3), 656–666.
- Kim, J., Lee, J., Li, X., Kunjravia, N., Rambhia, D., Cueto, I., Kim, K., Chaparala, V., Ko, Y., Garcet, S., Zhou, W., Cao, J., & Krueger, J. G. . Multi-omics segregate different transcriptomic impacts of anti-IL-17A blockade on type 17 T-cells and regulatory immune cells in psoriasis skin. Frontiers in Immunology (2023), 14, 1250504.
2022
- Kim, J., Lee, J., Hawkes, J. E., Li, X., Kunjravia, N., Rambhia, D., Cueto, I., Moreno, A., Hur, H., Garcet, S., Zhou, W., Cao, J., Krueger, J. G. (2022). Secukinumab improves mild-to-moderate psoriasis: A randomized, placebo-controlled exploratory clinical trial. Journal of the American Academy of Dermatology, 88(2), 428–430
- Qiu, C., Cao, J., Martin, B. K., Li, T., Welsh, I. C., Srivatsan, S., Huang, X., Calderon, D., Noble, W. S., Disteche, C. M., Murray, S. A., Spielmann, M., Moens, C. B., Trapnell, C., & Shendure, J. (2022). Systematic reconstruction of cellular trajectories across mouse embryogenesis. Nature Genetics, 54(3), 328–341.
- Martin, B. K., Qiu, C., Nichols, E., Phung, M., Green-Gladden, R., Srivatsan, S., Blecher-Gonen, R., Beliveau, B. J., Trapnell, C., Cao, J., Shendure, J. (2022). Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nature Protocols, 18(1), 188–207.
- Pattwell, S. S., Arora, S., Nuechterlein, N., Zager, M., Loeb, K. R., Cimino, P. J., Holland, N. C., Reche-Ley, N., Bolouri, H., Almiron Bonnin, D. A., Szulzewsky, F., Phadnis, V. V., Ozawa, T., Wagner, M. J., Haffner, M. C., Cao, J., Shendure, J., & Holland, E. C. (2022). Oncogenic role of a developmentally regulated splice variant. Science Advances, 8(40), eabo6789.
2021
- Zhou, W.,* Melamed, D.,* Banyai, G.,* Meyer, C., Tuschl, T., Wickens, M., Cao, J.#, Fields, S#. (2021). Expanding the binding specificity for RNA recognition by a PUF domain. Nature Communications, 12(1), 5107.
Featured publications Before 2021
- Junyue Cao, Diana R. O’Day, Hannah A. Pliner, Paul D. Kingsley, Mei Deng, Riza M. Daza, Michael A. Zager, Kimberly A. Aldinger, Ronnie Blecher, Fan Zhang, Malte Spielmann, James Palis, Dan Doherty, Frank J. Steemers, Ian A. Glass, Cole Trapnell#, Jay Shendure#. A human cell atlas of fetal gene expression Science. 2020 DOI: 10.1126/science.aba7721
- Junyue Cao#, Wei Zhou, Frank Steemers, Cole Trapnell, Jay Shendure#. Sci-fate characterizes the dynamics of gene expression in single cells Nature Biotechnology. 2020. https://doi.org/10.1038/s41587-020-0480-9
- Junyue Cao*, Malte Spielmann*, Xiaojie Qiu, Daniel M. Ibrahim, Xingfan Huang, Andrew J. Hill, Fan Zhang, Stefan Mundlos, Lena Christiansen, Frank J. Steemers, Cole Trapnell#, Jay Shendure#. The single cell transcriptional landscape of mammalian organogenesis. Nature. 2019, (7745):496-502
- Junyue Cao, Darren A Cusanovich, Vijay Ramani, Delasa Aghamirzaie, Hannah A Pliner, Andrew J Hill, Riza M Daza, Jose L McFaline-Figueroa, Jonathan S Packer, Lena Christiansen, Frank J Steemers, Andrew C Adey, Cole Trapnell#, Jay Shendure#. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018, 361(6409):1380-1385.
- Junyue Cao*, Jonathan S Packer*, Vijay Ramani, Darren A Cusanovich, Chau Huynh, Riza Daza, Xiaojie Qiu, Choli Lee, Scott N Furlan, Frank J Steemers, Andrew Adey, Robert H Waterston#, Cole Trapnell#, Jay Shendure#. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017, 357(6352):661-667.